In my day to day work, I use to extract data from PDF
files - especially reports. For a recent analysis, I had to work on data
extraction on PDFs from
WHO reports on meningitis.
I wanted to find an automated way to extract tables from multiple PDFs and work
on them to generate report using data-frames/tibble. A little search leads me to the tabulizer1 package which depends on
rJava. In this series of posts, I will show how I handled data wrangling just
after table extraction using tabulizer on one PDF report.
Packages I used and some overview of the work
- The
dplyrpackage for data wrangling. - The
tabulizerpackage for extracting table from PDFs. - The
DTpackage for just table formatting. - The
stringrpackage for working with string data. - The
purrrpackage for mapping functions.
library(dplyr)
library(stringr)
library(tabulizer)
library(DT)
library(purrr)Before we start, let’s take a look on the table we want to extract, just to figure out what is the final output we must have in mind. Here is an example of table on meningitis pathogens located on page 5 of the most recent report.
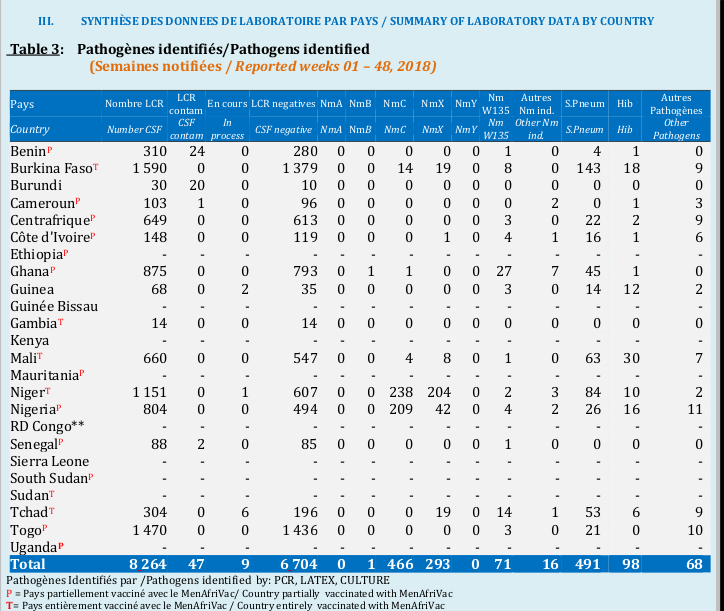
We will not go deeper in explaining the meaning of each column. We keep in mind that, at the end of our data management party -and it will be funny-, we want to have a kind of similar table, with these new column names that matches columns in the table presented above:
countries,number_csfcsf_contam,in_process,csf_negativenma,nmb,nmc,nmx,nmy,nmw135other_nm,s_pneumhibother_pathogens
Part one: extracting the row data
Once you succeed installing the tabulizer package, the extracting
part is fairly simple. The package has a function for it, extract_pdf
which has as arguments the file name, the page where we want to extract
the table, the desired method of extraction and the format we want to
get as result. Assuming my PDF is in the data folder we can write this:
pathogen_data <- extract_tables(
#the pdf file path
file = "data/meningitis-bulletin-s44-48-2018.pdf",
#the page where to extract the data. Remember, page 5.
page = 5,
#the output format
output = "data.frame",
#the method
method = "stream"
)About the output format
Once I launched the function extract_table, I thought we would have
as final output a data.frame of the table in page 5. But no.
class(pathogen_data)## [1] "list"The purpose behind is really understandable. If the page contains more than one table, all the tables of the page are compiled within a list where each table has the output format I told the function to hold in the parameters.
Also worth to mention, It would be really interesting to have a data-frame as output and not a matrix. Why? Because we are too busy to bother about pathogen_data[i+j-k, i-k+2] in the data
management part. Instead, we will focus on how to deal with problems
within our data-frame using the dplyr syntax. Take your broom and feel free to
add your inputs in comments.
Planning what to do.
Now let’s take a look on the data-frame (the only table) we have in the list. It’s its first element.
pathogen_final_data <- pathogen_data[[1]]
summary(pathogen_final_data)## X X.1 X.2
## Length:36 Length:36 Length:36
## Class :character Class :character Class :character
## Mode :character Mode :character Mode :characterA brief summary shows us only three variables in the data-frame, all characters
and named respectively X, X.1 and X.2. We might want to have a global look
on these three variables simultaneously, but I will move step by step since the data is really
a mess. Let’s create a function which will print just one selected column of my
data-frame pathogen_final_data. I want to access the column - which will be
passed as a parameter- of my data in a way that should be understood
by the select verb of dplyr, so I will put
tidy evaluation
in the game.
print_column <- function(.data, column, n_lines){
#enquo to quote the parameter
col <- enquo(column)
.data %>%
#bang bang to unquote
select(!!col) %>%
#print the number of lines
head(n = n_lines) %>%
datatable(class = "cell-border stripe",
option = list(
compact = TRUE,
pageLength = 6,
scrollX = TRUE),
rownames = FALSE)
}#It is a really mess, the first column
pathogen_final_data %>%
print_column(X, n_lines = 17)It seems that we have the countries are in the column named
X. This column has incorporated the section title and the name of the table. Also,
we have some T and P at the end of the country names and we might have some encoding
issues as you can see with one country Cote d'Ivoire in the table. There
is also some blank lines at the beginning of the table.
What about the second column?
#It is more messy the second column
pathogen_final_data %>%
print_column(X.1, n_lines = 12)Oops! We have a lot of empty lines at the beginning of the column. The columns of my final data-frame I have in mind as output seem to be merged together.
The third one?
pathogen_final_data %>%
print_column(X.2, n_lines = 13)OK. We even don’t know what is in this data-frame. Fortunately, We have the table’s look in the report. So here are what we are expected to do 2:
Split the columns so that we can have the data separated
Remove the empty lines at the top of the data-frame.
Remove the
TandPat the end of country names.Give meaningful column names to my data and convert to the correct type each column.
In this post, we are only going to focus on the first part.
Defining some little things
column names and country names
I am guilty about knowing the list of countries from where the data
has been taken. In order to be able to filter on those country names after,
we will create a vector named country_names with the name of the countries
which are covered.
We will also add the final name of columns we want to have at the end of
our data-wrangling party in the vector final_columns.
countries_names <- c('uganda', 'togo', 'tchad',
'sudan', 'south sudan', 'sierra leone',
'senegal', 'rd congo', 'nigeria',
'niger', 'mauritania', 'mali',
'kenya', 'gambia', 'guinee bissau',
'guinea', 'ghana', 'ethiopia',
"cote d'ivoire", 'centrafrique', 'cameroun',
'burundi', 'burkina faso', 'benin',
'tanzania')
#creating the final column heading
final_columns <- c("countries", "number_csf", "csf_contam",
"in_process", "csf_negative", "nma",
"nmb", "nmc", "nmx",
"nmy", "nmw35", "other_nm",
"s_pneum", "hib", "other_pathogens")Some little functions to start with
For tidying the data, I will need to remove some empty columns in the data-frame. I will also neet to work throughout filtering some columns where the percentage of empty values - or values with the dash “-” - is above a given threshold. Let’s work to define some functions for these purposes.
#find the percentage of empty values in a column
#vect is a character vector
percent_empty <- function(vect) sum(vect == ""| vect == "-") / length(vect)
#basically, the function can work with every data type, but let's consider
#only data-frame. The Threshold is exclusive
filter_empty_column <- function(.data, threshold = 1){
#filter in the data.frame, columns where the percentage
#of empty values is bellow the threshold.
Filter( function(x) (percent_empty(x) < threshold), .data)
}I need in my process to know exactly which column name I am indexing and be
sure about the column name. I created the rename_columns
function for that purpose.
The function will take a data-frame and change its
column names to column_1, column_2, …, column_n where n
is the number of columns of the data-frame.
#start will help indexing the begining of
#columns (either 0 or a given digit)
rename_columns <- function(.data, start = 0L){
#Getting the number of columns from the data-frame
nb_cols <- seq_along(colnames(.data)) + start
#Changing the column names
if( length(nb_cols) == 0L ){
#stop with empty data, to let me check where the error
#comes from
stop("no colnames found")
}
else{
colnames(.data) <- paste("columns", nb_cols, sep = "_")
#returning the data frame.
.data
}
}
#let'us take a look on an example dataset
mtcars %>%
as_tibble() %>%
rename_columns() %>%
print_column(everything(), n_line = 2)Splitting the columns and correcting the country names
Since the separator for merged columns is the space character, what will be interesting is to split every column in the data-frame. As we are planning to clean the country names , we will keep the first column when splitting.
#function for splitting a vector and returning a tibble
split_vector <- function(vect){
str_split(vect, "\\s", simplify = TRUE) %>%
as_tibble()
}
# a function to split the data
split_columns <- function(.data){
#first, renaming the columns of my data
.data <- rename_columns(.data)
#second, stock the first column somewhere
first_column <- .data %>%
select(columns_1)
#third, split all columns of the data.frame
splitted_data <- .data %>%
#using the purr package here
map_dfc(split_vector)
first_column
#finally, bind columns by including the first column
first_column %>%
bind_cols(splitted_data) %>%
#rename columns to be sure about the final output
rename_columns()
}In the writing process we will have to take a regular look on our data set. To avoid copy-pasting, we are going to create a little function to print the data-frame.
#function to print the pathogen_final_data
have_a_look <- function(){
pathogen_final_data %>%
print_column(columns_1:columns_10, n_lines = 10)
}
#splitting
pathogen_final_data <- split_columns(pathogen_final_data)
#printing
have_a_look()We learnt how to split all columns of a data-frame and keep the first column
within the final output and a use case of quotation.
In the next Step, we will deal with the three remaining problems.